It has been more than two years since the article “A state of flux” was published in the August 2015 issue of GCM. The article summarized the “known knowns” and “known unknowns” of off-type grasses in ultradwarf bermudagrass putting greens. The off-type issue continues to be a large part of the ultradwarf bermudagrass community, but research conducted since 2015 has aimed to answer some of these “known unknowns.” Some questions have been answered, and some remain.
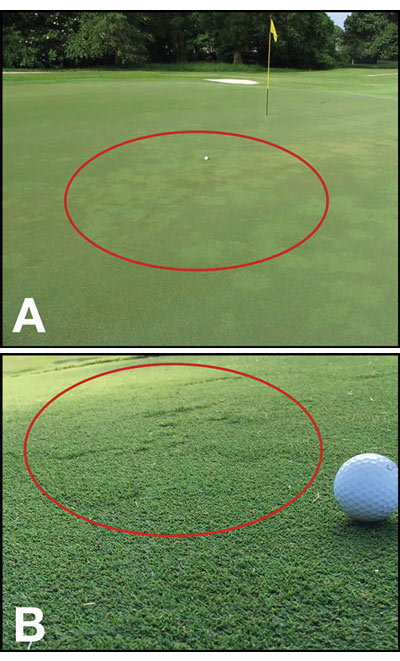
Right: Figure 1. Off-type grasses (lighter in color and noted by red circle) present in an ultradwarf bermudagrass putting green (A). Close-up of an off-type grass (noted by red circle) present in an ultradwarf bermudagrass putting green (B). Photos by Eric Reasor
Off-type grasses: What’s the issue?
Off-type grasses are defined as those with different morphology and performance compared with the desirable turfgrass. Figure 1 illustrates the aesthetic and surface uniformity disruptions the presence of off-types can cause in ultradwarf putting greens.
A genetic distinction does not have to exist for a grass to be considered an off-type. In fact, most of the genetic research has failed to distinguish ultradwarf cultivars Champion, MiniVerde and TifEagle from older cultivars such as Tifgreen and Tifdwarf despite vast differences in their morphology (6). This history of ultradwarf bermudagrass cultivars is much different than that of other bermudagrasses. The majority of cultivars used on putting greens were identified as off-type grasses in Tifgreen or Tifdwarf because of desirable morphological and performance differences.
The root cause of the off-type issue in the Tifgreen-derived cultivar family is still one of the “known unknowns.” A review article (6) has summarized the research of the genetic stability of Tifgreen and Tifdwarf, in addition to that of other hypothetical causes for off-types in ultradwarf bermudagrasses. Regardless of the cause or origin, off-type grasses are a major problem in ultradwarf putting greens. Research was conducted at the University of Tennessee from 2014 to 2017 to attempt to answer some of the concerns regarding this issue.
Building an off-type collection
The first step of this research was to sample off-type and desirable grasses on golf course putting greens. Beginning in summer 2013, 52 samples were harvested from 21 golf courses in Alabama, Arkansas, Florida, Mississippi, South Carolina and Tennessee. The superintendent at each golf course determined what was an off-type and what was the desirable grass.
It is important to note that off-type grasses were found in Champion, MiniVerde and TifEagle putting greens. A plug of each sample was taken to the University of Tennessee to be grown in a greenhouse environment for morphological and genetic characterization, as well as for responses to nitrogen and plant growth regulator (PGR) applications.
Morphological characterization
The off-type and desirable samples from putting greens were characterized according to their morphology. A single, three-node stolon of each sample was planted into a peat moss-based growing medium and maintained in a greenhouse environment. The stolon length at transplanting ranged from 1.3 to 4.4 inches (3.4 to 11.3 cm), which is an indication of the morphological and growth variability among the samples.
Morphological characterization of the off-type and desirable samples was conducted using methods similar to those used by other researchers (7). Internode length and stolon diameter were measured between the third and fourth node, and leaf length and width were quantified using the outer leaf from the third node. In addition, a leaf length:width ratio was calculated. All measurements were made using digital calipers, with each grass sample replicated four times and measurements collected on three stolons per replication (n = 12).
The morphological data were analyzed using a cluster analysis, with the goal of grouping these grasses according to the variability among samples. A clustering algorithm was used to separate the data set into a defined number of clusters (that is, groups). Three clusters were determined based on a clustering criterion number and the number of observations in each cluster.
Cluster 1 contained nine off-type and five desirable samples. Cluster 2 had 12 off-type and 14 desirable samples. Cluster 3 had eight off-type and four desirable samples. Means for each morphological measurement of the three clusters are presented in Figure 2.
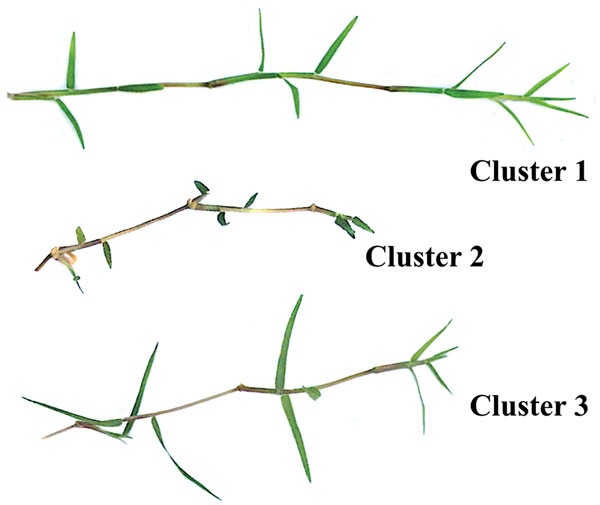
Figure 3. Photographs of samples representative of each morphological cluster. Grasses in Cluster 1 had significantly longer internode lengths than those in Clusters 2 and 3. Grasses in Cluster 3 had significantly longer leaves than those in Clusters 1 and 2.
Internode length, leaf length and length:width ratio were statistically different among the three clusters, whereas stolon diameter and leaf width were similar (Figure 2). The mean internode length for grasses in Cluster 1 was 0.5 inch (12.7 mm), and 0.4 inch (9.9 mm) longer than Cluster 2 or 3, respectively (Figure 2). However, leaf length was significantly longer in Cluster 3 (1.2 inches or 29.9 mm) than in Clusters 1 (0.6 inch or 14.9 mm) and 2 (0.4 inch or 9.9 mm). This relationship was also present in length:width ratio among clusters (Figure 2). Figure 3 (above) contains a representative sample from each morphological cluster.
Internode and leaf lengths varied greatly among desirable and off-type grasses as well as grasses measured in other experiments (4,7). This is an indication of the amount of phenotypic variability that can occur in an individual putting green, from golf course to golf course as well as cultivar to cultivar. Differences in internode and leaf length within the same putting surface can lead to decreased turfgrass density and reductions in putting surface quality and playability (6).
Genetic characterization
The inconsistencies of morphological measurements among hybrid bermudagrasses used on putting greens suggest genetic techniques may be more accurate in evaluating the diversity of these grasses. Several methods have been used to explore the genotypic differences among bermudagrass cultivars and off-type grasses. The methodology used in these previous experiments has been detailed elsewhere (6), but the majority of experiments share the same conclusion: Cultivars within the Tifgreen-derived family are not readily distinguished from one another using molecular marker technology (that is, DAF, AFLP and SSR).
One molecular marker method, genotyping-by-sequencing (GBS), had not been used to identify cultivars and off-type grasses. Genotyping-by-sequencing is capable of efficiently identifying large numbers of single nucleotide (DNA base) variants (1). In addition, genotyping-by-sequencing has been used with success in switchgrass (3), wheat and barley (5). Based on its robustness and its successful use in other grasses, we hypothesized that genotyping-by-sequencing might be able to identify genetic variation among off-type grasses and hybrid bermudagrasses used on putting greens.
DNA was isolated from bermudagrass samples and then sent to the Cornell University Institute for Biotechnology in Ithaca, N.Y., for genotyping-by-sequencing analysis. Samples of known Champion, MiniVerde, Tifdwarf, TifEagle, Tifgreen and Tifway were included as standards, along with 47 off-type and desirable grasses from putting greens. The initial step in GBS involved a restriction enzyme “cutting” the DNA into small pieces. Those small pieces of DNA were then fitted with barcode adapters for sequencing and sample recognition. After the small pieces of DNA were sequenced, the bioinformatics analysis pipeline sorted, indexed and merged the reads to determine nucleotide variants. Multidimensional scaling plots were then generated from these variants to illustrate the variation among samples (Figure 4).
The genotyping-by-sequencing results were surprising because the majority of samples harvested from golf courses clustered with the standard cultivars Champion, MiniVerde, Tifdwarf, TifEagle and Tifgreen in the multidimensional scaling plot (Figure 4). This clustering suggested that these samples were genetically similar to those cultivars. Only five (~11%) of the 47 unknown samples were genetically divergent from the standard cultivars. The ultradwarf cultivars were also genetically similar to Tifgreen and Tifdwarf, but genotyping-by-sequencing separated Tifway from the other hybrid cultivars. Our results using genotyping-by-sequencing were similar to previous molecular genetics research, which also failed to readily distinguish among ultradwarf cultivars and most off-type grasses.
So why did the majority of grasses included in our experiment (and others) exhibit variable morphological characteristics while being genetically similar? This is still a “known unknown” with the off-type issue. However, the majority of bermudagrass putting green cultivars were selected from other bermudagrass cultivars (6), and the off-type grasses studied here were also selected from existing plantings. The difference in morphology could be driven by differential gene expression influenced by environment or management practices (2,8). The intense management practices implemented on ultradwarf putting greens could result in the up- or down-regulation of genes that control important turfgrass characteristics (that is, internode and leaf length). However, no research has been conducted to explore this hypothesis.
Off-type responses to nitrogen and trinexapac-ethyl
We evaluated the response of off-type grasses and ultradwarf cultivars to nitrogen and trinexapac-ethyl applications. Three ultradwarf cultivars (Champion, MiniVerde and TifEagle) and three off-type grasses were treated with increasing nitrogen rates (0 to 1 pound nitrogen 1,000 square feet/week). One off-type was selected from each distinct morphological cluster. The grasses were established in greenhouse culture in a sand/peat mix meeting USGA root-zone recommendations. Daily clipping was suspended at the time of initial nitrogen treatment application, and growth above 1 cm (0.4 inch) was harvested every seven, 14, 21 and 28 days.
Pooled across all six grasses, the three lowest nitrogen treatments decreased weekly clipping production 17% to 29%, whereas the three highest nitrogen treatments sustained clipping production. The direct application of these results for putting green management is not clear because of the differences between plant culture in a greenhouse and that in the field. It is also expected that ultradwarf bermudagrasses maintained at 1 cm in a greenhouse would perform differently than plants maintained at putting green mowing heights. This was the first research on responses of off-type ultradwarfs to nitrogen applications, and field research should be carried out to support the results of this experiment.
In a separate experiment, trinexapac-ethyl (Primo MAXX, Syngenta) was applied to the same off-type grasses and ultradwarf cultivars in the greenhouse at 0, 0.1875, 0.375, 0.75, 1.5, 3, 6 or 12 fluid ounces/acre (13.7, 27.4, 54.8, 109.6, 219.2, 438.5 or 876.9 milliliter/hectare). On each date that significant differences were detected, mean clipping weights following application at 3 fluid ounces/acre were not significantly different from mean clipping weights following application at 6 or 12 fluid ounces/acre. These results suggest that, under the conditions of our experiment, there was not a benefit to applying rates greater than 3 fluid ounces/acre in a single application. However, field validation of this response is warranted.
Pooled across all Primo MAXX rates, the six grasses responded differently. Mean clipping weight for the off-type grass in cluster one (OTC1) was 21% to 27% greater than mean clipping weights for Champion, MiniVerde and off-types in clusters two and three. OTC1 (longer internode length) produced 15% more clippings than TifEagle in this experiment; however, this response was not statistically significant. The three off-type grasses and three commercial cultivars used in this experiment were genetically similar but exhibited differential responses to trinexapac-ethyl. Off-type grasses present in putting greens that are similar to OTC1 have the potential to disrupt the functional and aesthetic characteristics because of differential susceptibility to trinexapac-ethyl.
What can be done?
Regardless of the origin, morphology or genetics, off-type grasses continue to be an issue in ultradwarf putting greens. The management of putting greens with off-type grasses is still a “known unknown,” although we have researched a few beginning guidelines to help mitigate their presence.
The goal of managing these ultradwarf putting greens is to balance the growth of the desirable cultivar and the off-type. That being said, adjusting nitrogen application and PGR application programs (for example, trinexapac-ethyl) is critically important on ultradwarf putting greens with off-type grasses. Certain off-type grasses are less regulated by trinexapac-ethyl than desirable ultradwarf cultivars, and frequent applications (every three to seven days) at high rates (that is, > 3 fluid ounces/acre) can lead to severe growth suppression of the ultradwarf cultivar, whereas the off-type grass can still be actively growing. It is important to maintain consistent growth among morphologically different grasses in order to minimize any competitive growth advantage an off-type grass may possess over a desirable cultivar.
Future research will explore the management of putting greens with off-type infestations in field settings. One potential topic of interest is the use of growing degree-day models to schedule PGR applications. This approach has the potential to aid in balancing the amount of growth suppression between the ultradwarf cultivar and the off-type grass. In addition to application timings, other PGR active ingredients such as prohexadione-Ca should also be evaluated for their effectiveness in balancing growth suppression between desirable cultivars of ultradwarf bermudagrass and off-type grasses. While there are still many “known unknowns” regarding off-type grasses in ultradwarf bermudagrass putting greens, we hope that more research can begin to change these to “known knowns.”
Funding
The University of Tennessee Institute of Agriculture, Memphis Area GCSA and the United States Golf Association provided funding for this project.
Acknowledgments
The authors would like to thank Qi Sun, Jeff Dunne, Sarah Boggess, Anne Hatmaker, Monil Mehta, Laura Poplawski, Sujata Agarwal, Javier Vargas, Tyler Campbell, Jimmy Greenway, Greg Breeden, Daniel Farnsworth, Trevor Mills, Mitchell Riffey, Kelly Arnholt, Jason Burris, Amanda Webb, Rebecca Grantham, Gerald Henry, Brian Schwartz, Robert Trigiano, Margaret Staton, Phil Wadl, and John Sorochan for assistance with this project. Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement.
The research says ...
- The differences in morphology and performance between off-type grasses and ultradwarf bermudagrass disturb the aesthetics and surface uniformity of ultradwarf putting greens.
- Internode length, leaf length and leaf length:width ratio of 52 samples of off-type desirable grasses were measured to group the grasses by their morphology.
- Genotyping-by-sequencing was used to compare the genetic makeup of the grasses, and the results were similar to previous genetic research, which also failed to readily distinguish among ultradwarf cultivars and most off-type grasses.
- Two different greenhouse experiments tested the responses of the grasses to nitrogen or trinexapac-ethyl. The experiments need to be replicated in the field to validate the results.
Literature cited
- Elshire, R.J., J.C. Glaubitz, Q. Sun, J.A. Poland, K. Kawamoto, E.S. Buckler and S.E. Mitchell. 2011. A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLOS One 6:E19379. doi:10.1371/journal.pone.0019379
- Fei, S. 2008. Recent progresses on turfgrass molecular genetics and biotechnology. Horticulturae 783:247-260.
- Fiedler, J.D., C. Lanzatella, M. Okada, J. Jenkins, J. Schmutz and C.M. Tobias. 2012. High-density single nucleotide polymorphism linkage maps of lowland switchgrass using genotyping-by-sequencing. The Plant Genome 8(2). doi:10.3835/plantgenome2014.10.0065
- Magni, S., M. Gaetani, L. Caturegli, C. Leto, T. Tuttolomodo, S. La Bella, G. Virga, N. Ntoulas and M. Volterrani. 2014. Phenotypic traits and establishment speed of 44 turf bermudagrass accessions. Acta Agriculturae Scandinavica, Section B - Soil and Plant Science 64:722-733. doi:10.1080/09064710.2014.955524
- Poland, J.A., P.J. Brown, M.E. Sorrells and J.L. Jannink. 2012. Development of high-density genetic maps for barley and wheat using a novel two enzyme genotyping-by-sequencing approach. PLOS One 7:E32253. doi:10.1371/journal.pone.0032253
- Reasor, E.H., J.T. Brosnan, R.N. Trigiano, J.E. Elsner, G.M. Henry and B.M. Schwartz. 2016. The genetic and phenotypic variability of interspecific hybrid bermudagrasses (Cynodon dactylon (L.) Pers. × C. transvaalensis Burtt-Davy) used on golf course putting greens. Planta 244:761-773. doi:10.1007/s00425-016-2571-8
- Roche, M.B. and D.S. Loch. 2005. Morphological and development comparisons of seven greens quality hybrid bermudagrass [Cynodon dactylon (L.) Pers. × C. transvaalensis Burtt-Davy]. International Turfgrass Society Research Journal 10:627-634.
- Sahu, P.P., G. Pandey, N. Sharma, S. Puranik, M. Muthamilarason and M. Prasad. 2013. Epigenetic mechanisms of plant stress responses and adaptation. Plant Cell Reports 32:1151-1159. doi:10.1007/s00299-013-1462-x
-
Williams, S. 2015. A state of flux: Experts agree on the benefits of ultradwarf bermudagrasses, but are also paying heed to emerging challenges facing those who manage these warm-season turfgrasses. Golf Course Management 83(8):72-80.
Eric Reasor is an assistant professor in the Department of Plant and Soil Sciences, Mississippi State University, Mississippi State, and Jim Brosnan is an associate professor in the Department of Plant Sciences, University of Tennessee, Knoxville.