
Figure 1: Composite turf core samples of MS-Supreme ultradwarf bermudagrass collected from 26-square-foot (2.4-square-meter) areas of interest following core aerification in 2017 and 2018 at the Mississippi State University Golf Course in Starkville, Miss. Photo by Matthew A. Tucker
The motivation to pursue turfgrass pathology was triggered after attending a talk by Monica Elliott, Ph.D., on the etiology of bermudagrass decline at the 1993 International Turfgrass Society Conference (4). Her research identified Gaeumannomyces graminis
var. graminis (Gg), an ectotrophic root-infecting (ERI) fungus that attacked and rotted roots of bermudagrass putting greens. The disease was initially described as bermudagrass decline by Freeman and Augustin (5) in Florida. Irregularly shaped chlorotic
patches were observed along the margins of bermudagrass greens due to rotted roots. The foliar symptoms would appear in the late summer and early fall when temperatures and humidity were consistently high, and rainfall was frequent (2). Roots of affected
grass are short, black, brittle and dysfunctional (2).
Fast forward 20 years and hundreds of ERI fungal isolates later observed in the Turf Pathology Lab at Mississippi State University. We wanted to determine whether other ERI fungi were involved in bermudagrass decline, currently referred to as take-all
root rot (TARR). Six years of research and two graduate students later, we know additional ERI fungal species are associated with TARR (8, 9, 10). Our hypothesis was that a complex of ERI fungi may be associated with the root rot disease. Three novel
ERI species, Magnapothiopsis cynodontis (Mc), Gaeumannomyces nanograminis (Gn) and Candidacolonium cynodontis (Cc) were identified from symptomatic bermudagrass roots throughout the southeastern United States and Hawaii (1, 8, 9, 10). Research out
of Dr. Jim Kerns’ Turf Pathology Lab at North Carolina State University identified an additional ERI fungus, G. graminicola, infecting roots of bermudagrass greens in North Carolina (7).
With this new information, I and former Master of Science student, M. Aaron Tucker (current doctoral student under David McCall, Ph.D., Virginia Tech), aimed to better understand the extent of co-colonization of these ERI fungi on bermudagrass putting
green roots. We sampled two MS-Supreme ultradwarf bermudagrass greens at Mississippi State University Golf Course (MSUGC). One green was identified by Pat Sneed, CGCS, as healthy (no history of TARR) and one TARR-symptomatic. Our objectives were to
determine the frequency of occurrence (FO) and the inoculum density of selected ERI fungi associated with the roots. We utilized quantitative multiplex assays, developed in our lab, to rapidly identify the fungi of interest (1).
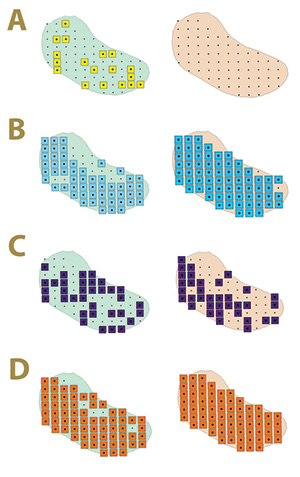
Figure 2. Fungal distribution of select ectotrophic root-infecting fungi within the Mississippi State University Golf Course Green 2 in 2017 (left) and 2018 (right), where A) yellow squares indicate areas positive for Gaeumannomyces graminis; B) blue squares indicate areas positive for G. nanograminis; C) purple squares indicate areas positive for Candidacolonium cynodontis; and D) orange squares indicate areas positive for Magnaporthiopsis cynodontis.
Materials and methods
Sampling methodology and root collection
A summer core aerification was conducted in July 2017 and 2018 on MS-Supreme Greens 2 (TARR asymptomatic) and 12 (TARR symptomatic) at MSUGC. Composite samples generated from turf cores provided root material for use in determining the presence of Gg,
Gn, Mc and Cc within designated areas of interest (AOI) on each green. Virtual maps were created using ArcGIS to establish AOIs for sampling by locating each associated latitude and longitude. A fishnet grid was added to each map to display AOIs on
9-foot (2.7-meter) centers within the footprint of each green. Shape files were created with each AOI assigned its latitude and longitude and transferred to a Trimble Geo 7x (Sunnydale, Calif.) handheld GPS unit. The center of each AOI was flagged
in both greens, which corresponded to the virtual map created through ArcGIS. Within a 26-square-foot (2.4-square-meter) grid, aerification cores were raked to the center to create a composite turf core sample for each AOI (Figure 1).
Roots were separated by hand from stolons and rinsed in a 2.5% sodium hexametaphosphate solution for 15 minutes to remove extraneous organic matter and sand particles. Root samples were blotted dry with sterile paper towels and stored at 39 F (4 C) for
subsequent DNA extraction.
Identification, distribution and inoculum density of select ERI fungi
Total genomic DNA (gDNA) was extracted from each composite root sample. Real time PCR (qPCR) was performed with the gDNA extracted from each root sample. Two multiplex assays were designed to specifically identify Gg and Gn (Assay I) and Mc and Cc (Assay
II). Each assay also was designed to identify bermudagrass root material. These assays allowed intensive root sampling on the greens and replaced the traditional pure culture fungal isolation and identification from roots.
Results from qPCR multiplex assays were used to determine the frequency of occurrence for each fungus within a green in 2017 and 2018. The inoculum density (number of DNA copies) of select ERI fungi in a green within a year and the inoculum density of
each ERI fungus within a green across years were completely randomized with three subsamples per composite root sample and each gDNA sample duplicated in the multiplex Assays I and II. These data were subjected to the analysis of variance using PROC
GLM of SAS. Fisher’s protected least significant difference test was used when means were significant (P≤0.05).
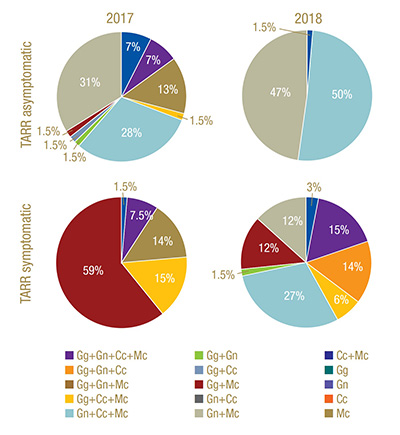
Figure 3. Frequency of occurrence of select ectotrophic root-infecting fungi complexes identified in Greens 2 (TARR asymptomatic) and 12 (TARR symptomatic) at Mississippi State University Golf Course in 2017 and 2018. TARR=take-all root rot; Gg=Gaeumannomyces graminis; Gn=G. nanograminis; Cc=Candidacolonium cynodontis; Mc=Magnaporthiopsis cynodontis.
Results
Sampling methodology and root collection
Green 2 (TARR asymptomatic) consisted of 68 AOIs, and Green 12 (TARR symptomatic) had 66 AOIs. The location of AOIs for each green remained consistent in 2017 and 2018. The mean composite turf core sample for each AOI was 1 pound (454 grams). Turf core
sampling and analysis were repeated in 2018. To summarize, Greens 2 and 12 had in total 864 and 816 experimental units, respectively, for the two-year study. The composite root samples ranged in appearance from healthy (i.e., white, fibrous roots)
to dark, necrotic, brittle roots.
Identification, distribution and inoculum density of select ERI fungi
Green 2 (TARR asymptomatic)
Sixty-seven of 68 AOIs in Green 2 had roots colonized by at least one ERI fungus in 2017, and roots collected from all 68 AOIs were colonized in 2018. The frequency of occurrence (FO) of Gg was 26%; however, there was a 100% reduction in FO in 2018. In
contrast, the FO for Gn was 97%, an increase of 20% from 2017 to 2018. A slight 4% increase in Mc FO was observed from 90% to 100% FO in 2017 and 2018, respectively. Cc had an FO of 50% and 51%, remaining steady across both years. The FO of the ERI
fungi was not static from 2017 to 2018 in Green 2, nor was their distribution within the green. The colonization of bermudagrass roots by the ERI fungi changed within AOIs across years (Figure 2). A complex of two or more ERI fungi was also identified
in 92% and 99% of AOIs in 2017 and 2018, respectively. Gn and Mc had the highest FO for co-colonization of roots in 2017, and Gn, Mc and Cc co-colonized roots at the highest FO in 2018 in Green 2. A four-ERI fungal complex was also observed in 2017.
(Figure 3).
The inoculum density of select ERI fungi was based on the number of DNA copies extracted from the bermudagrass roots and determined in multiplex Assays I and II. The main effect, fungus, influenced inoculum density in Greens 2 and 12. The inoculum density
of the ERI fungi in the TARR asymptomatic Green 2 ranged from 123 to 59,368 DNA copies in 2017; however, no differences were observed among the ERI fungi (P=0.0994). In 2018, inoculum density in Green 2 ranged from 0 to 142,947. Gg was not identified
in the green in 2018 but had similar inoculum density to that of Mc (P=0.0374). Gn had the greatest inoculum density, 70% more than Cc (Table 1). The inoculum density of each ERI fungus identified in Green 2 fluctuated from 2017 to 2018, but only
Mc had significant changes in inoculum density (P<.0001) with a 74% increase from 2017 to 2018 (Table 2).
Green 12 (TARR symptomatic)
At least one select ERI fungus was identified in the roots of all 66 AOIs within Green 12 in 2017, and only one AOI did not have root colonization in 2018. Gg, originally reported as the etiological agent of bermudagrass decline (TARR) (3), resulted in
a 95% FO in 2017 in contrast to 48% FO in 2018; a 49% reduction in root colonization. The FO of Gn was only 20% in 2017 but increased to 70% in 2018. The ubiquitous Mc was identified in the majority of root samples in both years with FO of 98% and
97% in 2017 and 2018, respectively. The FO of Cc increased 53% from 2017 to 2018, resulting in an FO of 51%. As observed in Green 2, the select ERI fungal colonization of bermudagrass roots varied within Green 12 in the two-year study (Figure 4).
Co-colonization with at least two ERI fungi was present in 97% of the AOIs in 2017. Gg and Mc were most frequently co-colonizing bermudagrass roots in 2017. A co-colonization of all four ERI fungi was observed at a low frequency in 2017 but increased
100% in 2018. Mc was a co-colonizing partner with the other three ERI fungi in 2017 (Figure 3).
The inoculum density of Gg was 81 times greater or more than that of the other ERI fungi present in Green 12 (Table 1). Although Mc had the lowest inoculum density in 2017, it was similar to Gn and Cc (P=0.0050). No differences in inoculum density were
observed among ERI fungi in 2018 (P=0.3972), but again, Mc had numerically the lowest inoculum density (Table 1). Inoculum density was affected when comparing fungus across years. Gg had 26 times greater inoculum density in 2017 than 2018. Despite
low inoculum density values for Mc in both years, 2018 resulted in a fivefold increase. The inoculum density of Gn and Cc remained similar across years (P=0.2404 and P=0.4152, respectively) (Table 2).
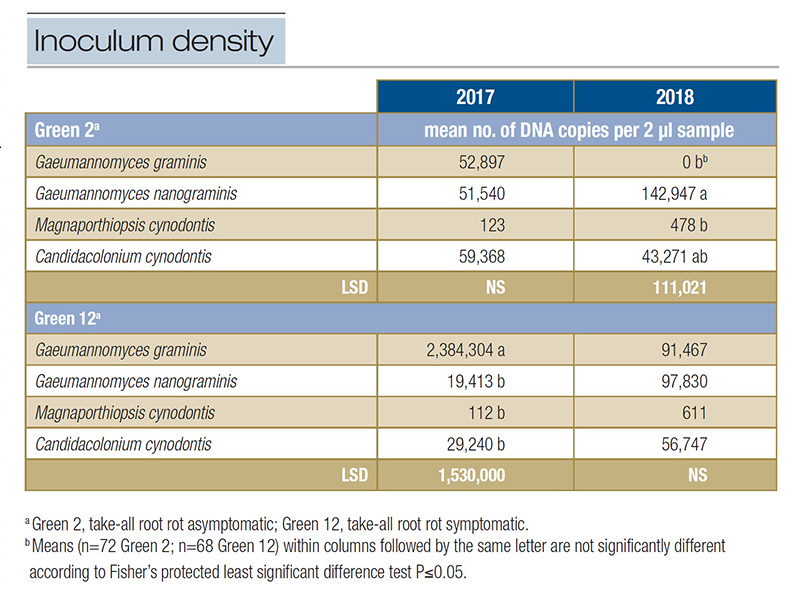
Table 1. Inoculum density of select ectotrophic root-infecting fungi present within two greens at Mississippi State University Golf Course in 2017 and 2018.
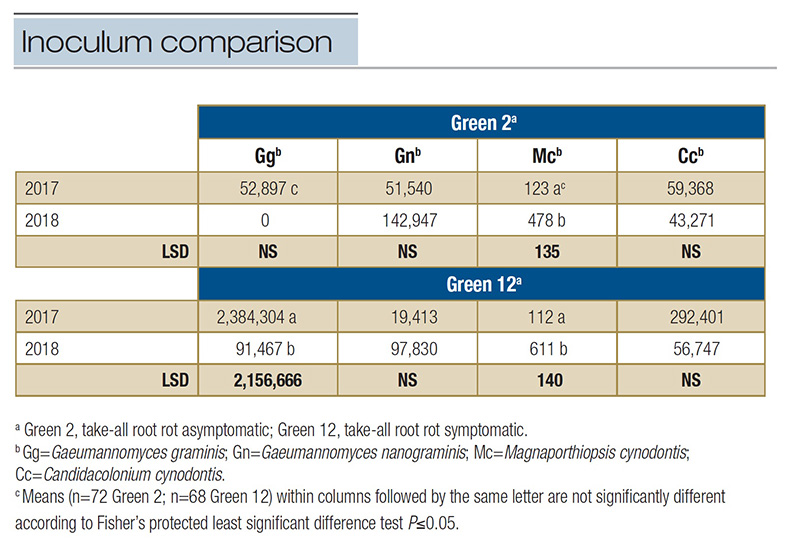
Table 2. Comparison across years of the inoculum densities of select ectotrophic root-infecting fungi identified in two greens at Mississippi State University Golf Course in 2017 and 2018. Inoculum density represents the mean number of DNA copies per 2 µl sample within a green each year analyzed from qPCR multiplex assays specific to each ERI fungus.
Conclusions
The results of this research were successful in identifying select ERI fungi widely distributed throughout two bermudagrass greens at a daily operational golf course using intensive sampling and qPCR multiplex assays. More commonly, two or more ERI fungi
formed a complex, co-colonizing bermudagrass roots. These findings are supported by results of Bronzato-Badial et al. (1) and most recently by Stephens et al. (6). Prior to this study, personal observations of bermudagrass samples revealed ERI fungi
other than Gg were associated with root rot leading to decline or from symptomatic TARR patches. Many times, ERI fungi were observed on rotted roots while the turf canopy was asymptomatic. We demonstrated in this study that Green 2 appeared asymptomatic
while roots were colonized with multiple ERI fungi.
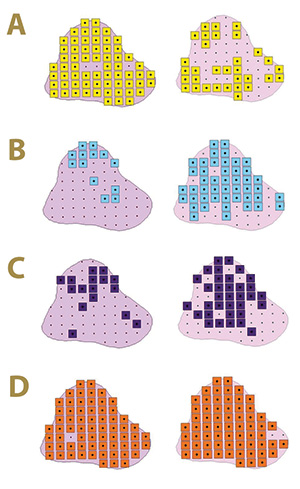
Figure 4. Fungal distribution of select ectotrophic root-infecting fungi within the Mississippi State University Golf Course Green 12 in 2017 (left) and 2018 (right), where A) yellow squares indicate areas positive for Gaeumannomyces graminis; B) blue squares indicate areas positive for G. nanograminis; C) purple squares indicate areas positive for Candidacolonium cynodontis; and D) orange squares indicate areas positive for Magnaporthiopsis cynodontis.
The distribution of select ERI fungi in Greens 2 and 12 was not fixed across years. This may indicate these fungi are resident in the thatch layer throughout the greens, and when new roots are produced, the possibility of colonization by one or more ERI
fungi is likely. Less than 6% AOIs were negative for root colonization by the select ERI fungi in this study. This supports the importance for golf course superintendents managing bermudagrass greens to minimize plant stress and provide optimal growing
conditions and inputs to maintain healthy greens. Monitoring plant health and understanding the distribution of these ERI fungi within bermudagrass greens will heighten the awareness of golf course superintendents to be prepared for potential root
rot and TARR symptoms within greens.
The success of this research was attributed to the application of locked nucleic acid probe technology, which maximized specificity and ERI fungal identity from root material. This technology can be implemented as a rapid identification tool of ERI fungi
in plant diagnostic laboratories receiving turfgrass samples. Within the last decade, research on ERI fungi associated with warm-season turfgrass species has increased our understanding and knowledge base. Continued efforts should be made to better
understand the biology of ERI fungi to in turn make knowledge-based management decisions to reduce root rot and decline of turf.
Funding
Mississippi Agricultural and Forestry Experiment Station, Mississippi State University and the National Institute of Food and Agriculture, U.S. Department of Agriculture, Hatch project under accession number 213130; Syngenta, BASF, BayerCrop Science,
PBI-Gordon and Helena.
The research says ...
- Gaeumannomyces graminis is purported to be the causal agent of take-all root rot of bermudagrass putting greens.
- Ectotrophic root-infecting fungal complexes colonize bermudagrass roots.
- Magnaporthiopsis cynodontis was the predominant fungus identified in bermudagrass roots.
- Gaeumannomyces nanograminis colonized bermudagrass roots more often than G. graminis in healthy roots
- Geaumannomyces graminis colonized bermudagrass roots more often than G. nanograminis in a declining green.
Acknowledgements
This research was conducted by M. Aaron Tucker to satisfy the research component of his Master of Science in Turfgrass Pathology in the Department of Biochemistry, Molecular Biology, Entomology and Plant Pathology, Mississippi State University.
Thank you to Pat Sneed, CGCS, Mississippi State University Golf Course, and Jason Ruffin, turfgrass manager, Mississippi State University Turfgrass Research Center, for their assistance and cooperation in this research.
Literature cited
- Bronzato-Badial, A., J. King and M. Tomaso-Peterson. 2020. Monitoring ectotrophic root-infecting fungi associated with bermudagrass putting green roots using quantitative multiplex assays. Plant Health Progress 21(2):144–151 (https://doi.org/10.1094/PHP-02-20-0010-RS).
- Elliott, M.L., and P.J. Landschoot. 1991. Fungi similar to Gaeumannomyces associated with root rot of turfgrasses in Florida. Plant Disease 75:238–241.
- Elliott, M.L. 1991. Determination of an etiological agent of bermudagrass decline. Phytopathology 81(11):1380–1384.
- Elliott, M.L. 1993. Bermudagrass decline: Transmission of the causal agent Gaeumannomyces graminis var. graminis by vegetative planting material. International Turfgrass Society Research Journal 7:329-334.
- Freeman, T.E., and B.J. Augustin. 1984. Bermudagrass decline. Florida Cooperative Extension Service, Institute of Food and Agricultural Sciences, University of Florida.
- Stephens, C., T.W. Gannon, M. Cubeta, T. Sit and J. Kerns. 2022. Characterization and aggressiveness of take-all root rot pathogens isolated from symptomatic bermudagrass putting greens. Phytopathology (https://doi.org/10.1094/PHYTO-05-21-0215-R).
- Stephens, C.M., and J.P. Kerns. 2020. First report of Gaeumannomyces graminicola causing bermudagrass decline of ultradwarf bermudagrass putting greens in North Carolina. Plant Disease 104(5):1552 (https://doi.org/10.1094/PDIS-10-19-2147-PDN).
- Vines, P.L., F.G. Hoffman, F. Meyer, T.W. Allen, J. Lou, N. Zhang and M. Tomaso-Peterson. 2020. Magnaporthiopsis cynodontis, a novel turfgrass pathogen with widespread distribution in the United States. Mycologia 112(1):52–63 (https://doi.org/10.1080/00275514.2019.1676614).
- Vines, P.L., F.G. Hoffmann, F. Meyer, T.W. Allen and M. Tomaso-Peterson. 2021. Gaeumannomyces nanograminis, sp. nov., a hyphopodiate fungus identified from diseased roots of ultradwarf bermudagrass in the United States. Mycologia 113(5):938–948
(https://doi.org/10.1080/00275514.2021.1911192).
- Vines, P.L., and M. Tomaso-Peterson. 2021. Candidacolonium cynodontis P.L. Vines and M. Tomaso-Peterson, gen. et sp. nov. Index Fungorum (http://indexfungorum.org/Names/NamesRecord.asp?RecordID=550880).
Maria Tomaso-Peterson (MariaT@pss.msstate.edu) is professor emerita, Matthew A. Tucker is a former graduate research assistant and Aline Bronzato-Badial is a former research associate, all in the Department of Biochemistry, Molecular Biology, Entomology
& Plant Pathology at Mississippi State University, Starkville, Miss.; James D. McCurdy is an associate professor and turfgrass Extension specialist in the Department of Plant and Soil Sciences, Mississippi State University; and Phillip L. Vines
is an assistant research professor in the Department of Plant Biology, Rutgers University, New Brunswick, N.J.