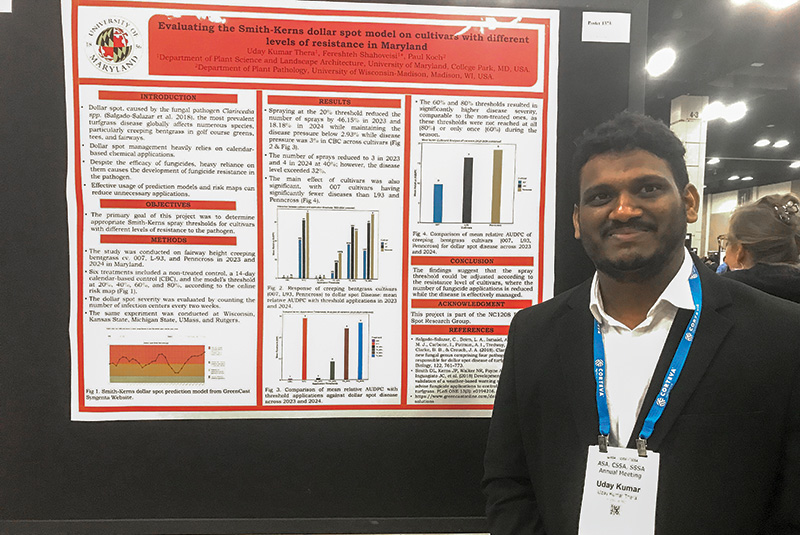
Evaluating the Smith-Kerns dollar spot model on cultivars with different levels of resistance in Maryland
Dollar spot, caused by the fungal pathogen Clarireedia spp., is the most prevalent turfgrass disease globally, affecting numerous species, particularly creeping bentgrass, in golf course greens, tees and fairways. Dollar spot management heavily relies on calendar-based chemical applications. Weather-based disease warning systems, such as the Smith-Kerns model, have been developed to enhance the timing of fungicide applications. However, further research is required to validate the model’s threshold on cultivars with different levels of disease resistance. A multistate NC1208 dollar spot research project was designed to test the Smith-Kerns model threshold on several creeping bentgrass cultivars. Two years of study in Maryland was conducted on fairway-height creeping bentgrass cultivars 007, L-93 and Penncross. Six treatments included a non-treated control, a 14-day calendar-based application and the model’s threshold at 20%, 40%, 60% and 80%. The dollar spot severity was evaluated by counting the number of infection centers.
The results indicated that using the 20% threshold led to seven spray applications in the 2023 season and nine in 2024, whereas the calendar-based approach resulted in 13 applications in 2023 and 11 in 2024. The relative area under the disease progress curve (AUDPC) at the 20% threshold across all cultivars was 0.848, while the relative-AUDPC for calendar-based sprays was 0.869. However, this difference was not statistically significant. Four sprays at the 40% threshold were required according to the model. This threshold level provided acceptable results in the semi-resistant cultivar 007 and statistically higher disease pressure in more susceptible cultivars. The threshold levels of 60% and 80% did not provide acceptable disease controls in any cultivars compared to the non-treated plots. These preliminary findings suggest that validating prediction models on cultivars with varying levels of disease resistance enhances the reliability of warning systems for turfgrass practitioners.
— Uday Kumar Thera (uthera@umd.edu), and Fereshteh Shahoveisi, Ph.D., University of Maryland, College Park, and Paul L. Koch, Ph.D., University of Wisconsin-Madison
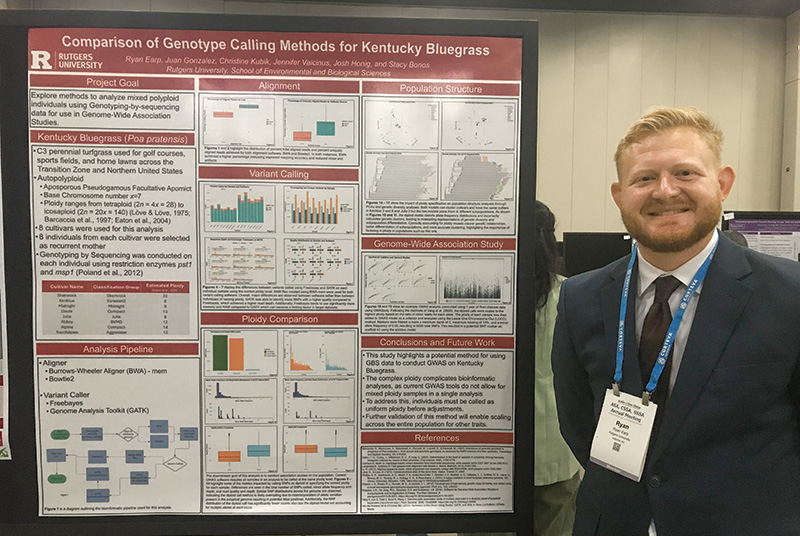
Comparison of genotype calling methods for Kentucky bluegrass (Poa pratensis)
Kentucky bluegrass (Poa pratensis L.) is a cool-season perennial turfgrass widely used across the U.S. and Canada, particularly on sports fields and golf courses (Huff, 2010). Performing genomic analyses has been difficult due to its apomictic reproduction, polyploidy and large genome size. However, the recent publication of a reference genome by Phillips et al. (2023) has helped reduce this complexity. This study evaluates methods for analyzing genotyping-by-sequencing data to perform a genome-wide association study (GWAS) on a Kentucky bluegrass population. A population of 62 individuals from eight cultivars was analyzed using two alignment tools, BWA and Bowtie2, and two variant callers, Freebayes and GATK.
BWA was able to align a higher percentage of total and unique reads to the reference genome compared to Bowtie2. GATK identified more variants with higher quality, while Freebayes showed higher read depth. Since GWAS software cannot handle mixed ploidy, GATK calls were compared using both uniform diploid and respective ploidy approaches. The diploid method resulted in 2,130,151 SNPs, with a median quality of 192 and depth of 48, while the polyploid method yielded 1,148,481 SNPs, with median quality of 551 and depth of 30. Both methods were able to cluster cultivars together using PCA and genetic diversity trees. A GWAS on rust disease data, collected in 2023, was conducted using the uniform diploid call, scaling to the highest sample ploidy, and incorporating ploidy as a cofactor in GWASpoly, following Yang et al. (2020). After filtering, 6,698 SNPs were used, identifying a potential SNP on scaffold 43. This study outlines a potential method for GWAS in mixed ploidy Kentucky bluegrass populations. Further research is needed to refine the model for larger populations and more traits to assist in improving breeding selection.
— Ryan Earp (ree43@sebs.rutgers.edu), Jennifer Vaiciunas, Christine Kubik, Josh A. Honig, Ph.D., and Stacy A. Bonos, Ph.D., Rutgers University, New Brunswick, N.J.
Darrell J. Pehr (dpehr@gcsaa.org) is GCM’s science editor.